chemfp performance
Chemfp is fast. As far as I can tell, chemfp is the fastest single-threaded similarity search program. Even on a laptop it can process around 100 million fingerprints per second.
As I've found out, detailed timings like the numbers below aren't all that useful because chemfp reached the point where overall performance is no longer based on the CPU, which are so fast that the limiting factor for single-threaded search is memory bandwidth. The computer architecture plays a more critical role when computing a sparse similarity matrix, because of the interplay of CPU type, multiple threads, BitBound limits, and processor caches. Even something as seemingly trivial as processing queries in popcount order improves performance.
The best solution is to request a license key to evaluate chemfp's performance yourself. Back in 2018 I generated a range of timing numbers for chemfp 3.2.1. Even if the hardware is now years out of date, it may still give you an idea of what search performance is like.
The results here were generated using the chemfp_benchmark. It uses test data sets with 166-bit (OpenEye MACCS), 881-bit (PubChem), 1024-bit (Open Babel FP2), and 2048-bit (RDKit Morgan) fingerprints. The 881-bit fingerprints were extracted from randomly selected PubChem records and the others were generated using randomly selected ChEMBL records.
Each target data set contains 1 million fingerprints. The corresponding query data set is not a subset of the target data set but is instead drawn from the full set of input fingerprints.
The benchmark then runs a number of different operations to get a sense of the performance.
The benchmark ran on machine with an "Intel Core i7-4770 CPU @ 3.40GHz" running at 3.7 GHz using Python 3.6.2 and chemfp 3.2.1.
Chemfp currently uses a single-threaded search for each query fingerprint, so the single query searches are not affected by the number of processors. In general the performance is linear in the number of fingerprints, so a 100M fingerprint database should take 100x longer than the times reported here.
k=10, single fingerprint query, in-memory search
Chemfp is designed to be called directly from a web application, assuming that application is written in Python. For example, you might have an application which displays information about a structure, and you want to change it to also display the nearest 10 neighbors in a target dataset containing a few million fingerprints. In that case you want the single-threaded search performance to be fast enough that it doesn't add noticeable overhead to the overall response time, even in the worst case.
This benchmark reports the timings of 1,000 k=10 nearest-neighbor queries for each of the 1M target data sets. In general the performance should be linear in the number of bits. The graphs below show that the fingerprint type also affects the performance.
It is also possible to ask chemfp to find the k-nearest neighbors with a similarity at or above a given threshold, which would reduce the search time.
| #bits | average (in ms) |
median (in ms) |
max (in ms) |
histogram |
|---|---|---|---|---|
| 166 | 1.1 | 1.0 | 3.4 | 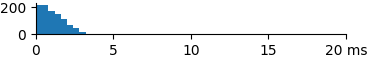 |
| 881 | 1.9 | 1.7 | 7.4 | 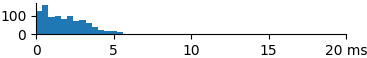 |
| 1024 | 3.3 | 3.0 | 9.6 |  |
| 2048 | 16.0 | 17.0 | 19.0 | 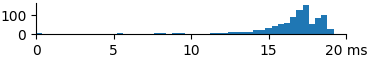 |
k-nearest scaling, single fingerprint query, 1024 bits, in-memory search
A k=1 nearest search is faster than a k=10 or k=100 nearest search. The following benchmark shows the effect of changing the value of k for the 1024-bit data set.
| k | average (in ms) |
median (in ms) |
max (in ms) |
histogram |
|---|---|---|---|---|
| 1 | 1.23 | 0.62 | 9.2 | 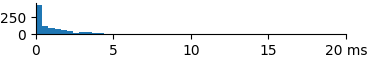 |
| 10 | 3.3 | 3.0 | 9.6 |  |
| 100 | 5.7 | 6.2 | 9.9 |  |
| 1000 | 8.1 | 9.3 | 11.0 |  |
threshold=0.7, single fingerprint query, in-memory search
This benchmark finds all matches at or above 0.7, and not just the first k. This is a high threshold, which means chemfp's sublinear search bounds are able to speed up the search by skipping most of the search space.
| #bits | average (in ms) |
median (in ms) |
max (in ms) |
histogram |
|---|---|---|---|---|
| 166 | 1.7 | 2.0 | 2.6 | 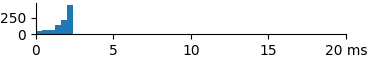 |
| 881 | 4.1 | 4.8 | 7.3 | 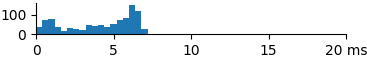 |
| 1024 | 4.6 | 5.2 | 6.6 | 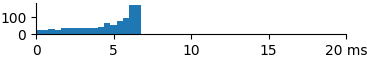 |
| 2048 | 12.0 | 14.7 | 14.4 |  |
NxN clustering
The previous benchmarks show the timing of a single query against a data set. Chemfp also supports "NxM" search to find similar matches for multiple queries against the data base. The NxM search is parallelized using OpenMP, where each query is run in its own thread.
Chemfp also has special support for the "NxN" case, where the same data set is used as both the queries and targets, excluding the diagonal. This is often used to construct a similarity matrix for clustering, along with the optimization that any similarity below a certain threshold can be treated as having no similarity.
In general, the similarity matrix construction time for the NxN case grows as N2 in the number of elements, linear in the number of fingerprint bits, and roughly linear in the number of processors until the memory bandwidth is saturated.
That is, each core is pulling data out of main memory as fast as it can. Let's say it processes 1Gbit per second, and the main memory bandwidth is 20Gbit/s. Each core adds another 1GBit per second, so with 20 CPUs there is simply no way for the memory to get to the CPU. This model is overly simple, but it helps understand why doubling the number of CPUs doesn't always double the performance.
In practice there is decent speedup on a 15-core machine. No one has reported their experience on larger configurations.
The following benchmarks try to convey an idea of how chemfp scales with the number of processors, fingerprint size/type, and threshold value. For the 250K and 500K cases, the fingerprint subsets were randomly sampled and the timings run several times; the timing variability was at the level of the least significant digits.
| #bits | #processors | average time (in s) |
scaleup |
|---|---|---|---|
| 166 | 1 | 70 | 1 |
| 166 | 2 | 37 | 1.9 |
| 166 | 4 | 21 | 3.2 |
| 881 | 1 | 166 | 1 |
| 881 | 2 | 93 | 1.8 |
| 881 | 4 | 54 | 3.1 |
| 1024 | 1 | 160. | 1 |
| 1024 | 2 | 82 | 1.9 |
| 1024 | 4 | 42 | 3.8 |
| 2048 | 1 | 362 | 1 |
| 2048 | 2 | 207 | 1.8 |
| 2048 | 4 | 102 | 3.5 |
| #bits | threshold | average time (in s) |
|---|---|---|
| 166 | 0.99 | 1.9 |
| 166 | 0.95 | 3.7 |
| 166 | 0.90 | 6.1 |
| 166 | 0.80 | 11 |
| 166 | 0.70 | 23 |
| 1024 | 0.99 | 2.5 |
| 1024 | 0.95 | 7.0 |
| 1024 | 0.90 | 12.8 |
| 1024 | 0.80 | 25.0 |
| 1024 | 0.70 | 42.4 |
| 2048 | 0.99 | 7.9 |
| 2048 | 0.95 | 17.5 |
| 2048 | 0.90 | 37 |
| 2048 | 0.80 | 72 |
| 2048 | 0.70 | 103 |
| #bits | #fingerprints | time (in s) |
time/time250,000 |
|---|---|---|---|
| 166 | 250,000 | 11 | 1 |
| 166 | 500,000 | 43 | 3.9 |
| 166 | 1,000,000 | 174 (= 2m 54s) |
15.8 |
| 1024 | 250,000 | 25 | 1 |
| 1024 | 500,000 | 119 | 4.8 |
| 1024 | 1,000,000 | 504 (= 8m 24s) |
20. |
| 2048 | 250,000 | 72 | 1 |
| 2048 | 500,000 | 312 (= 5m 12s) |
4.3 |
| 2048 | 1,000,000 | 1360 (= 22m 40s) |
19 |